Ocean Genomics, pioneers in AI-driven transcriptome analysis and biomarker discovery, announces the following abstract performed in collaboration with Aichi Medical Center, Nagoya, the National Cancer Center Hospital East, Kashiwa, and other leading centers in Japan.
“Discovery of a potential predictive marker for eribulin treatment and novel target genes in BRAF V600E mutant metastatic colorectal cancer using an AI-driven RNA-seq analysis platform: Translational research of the BRAVERY study (EPOC1701)”
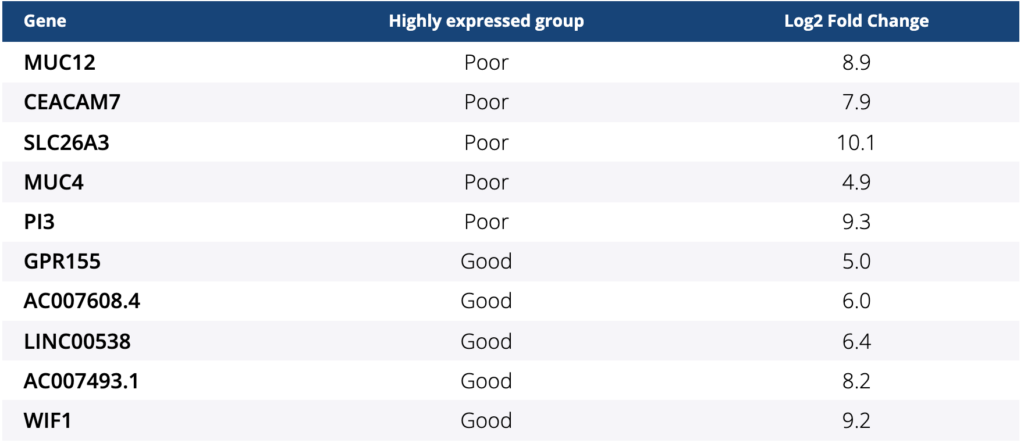
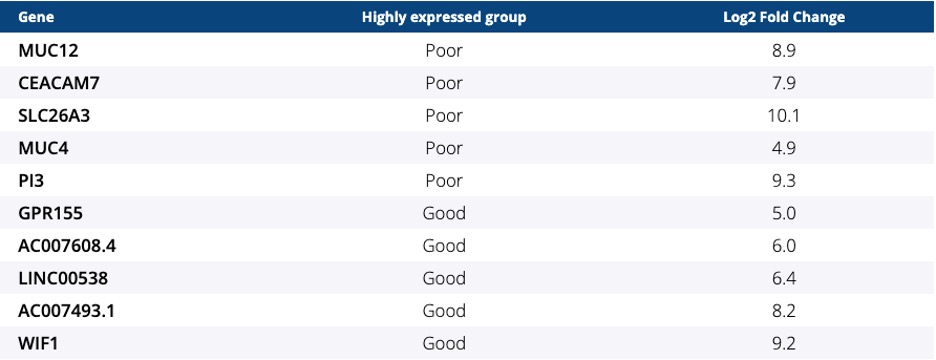
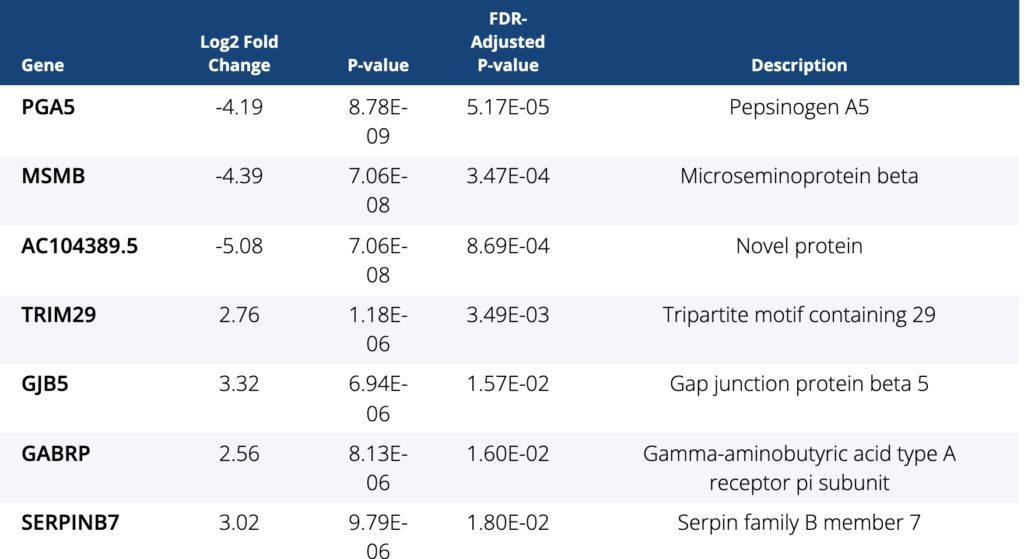
Ocean Genomics leveraged its AI-driven transcriptome analysis and biomarker discovery platform, txome.ai, to perform the analysis of a metastatic colorectal cancer cohort using RNA sequencing FASTQ files and matched clinical data supplied by the centers in Japan from the BRAVERY trial.
The abstract concluded, “The gene expression analyses suggest that BM2 subtype could be a predictive marker for the efficacy of eribulin and some genes could be novel targets with the goal to improve prognosis of pts with BRAF V600E mt mCRC. This is the first finding for a potential biomarker in this subgroup using RNA-seq analysis tools. These findings will require additional validation.”
The abstract can be found here: https://meetinglibrary.asco.org/record/198553/abstract.
Dr. Takayuki Yoshino, one of the principal investigators, noted, “This was the first and an important collaboration step to address the emerging field of biomarker discovery using RNA-seq when combined with A.I.-driven tools such as those from Ocean Genomics. We will aggressively move forward to a further verification using additional cohorts in the near future, together with Ocean Genomics”.
Dr. Takayuki Yoshino, M.D., PhD., currently works at the National Cancer Center Hospital East (NCCHE) in Chiba, Japan, where he is the Director for the Department of Gastroenterology and Gastrointestinal Oncology and the Head of the Clinical Research Coordinating Division.
“The results of this study are encouraging and highlight the opportunity to improve treatment selection using RNA-seq in biomarker development,” said Carl Kingsford, CEO of Ocean Genomics, Inc. “We look forward to validation and continued collaboration with Dr. Yoshino and the outstanding teams at NCCHE, Aichi Medical Center and affiliated centers of excellence.”
For further information on Ocean Genomics and our txome.ai platform:
Please visit our website: https://oceangenomics.com
Or email us: contact@oceangenomics.com